Pairwise registration for grouped cardiac MR images¶
Note: Please read the DeepReg Demo Disclaimer.
This demo uses the grouped dataset loader to register intra-subject multi-sequence cardiac magnetic resonance (CMR) images.
Author¶
DeepReg Development Team
Application¶
Computer-assisted management for patients suffering from myocardial infraction (MI) often requires quantifying the difference and comprising the multiple sequences, such as the late gadolinium enhancement (LGE) CMR sequence MI, the T2-weighted CMR. They collectively provide radiological information otherwise unavailable during clinical practice.
Data¶
This demo uses CMR images from 45 patients, acquired from the MyoPS2020 challenge held in conjunction with MICCAI 2020.
Instruction¶
Installation¶
Please install DeepReg following the instructions and
change the current directory to the root directory of DeepReg project, i.e. DeepReg/.
Download data¶
Please execute the following command to download/pre-process the data and download the pre-trained model. Images are re-sampled to an isotropic voxel size.
python demos/grouped_mr_heart/demo_data.py
Launch demo training¶
Please execute the following command to launch a demo training (the first of the ten
runs of a 9-fold cross-validation). The training logs and model checkpoints will be
saved under demos/grouped_mr_heart/logs_train.
python demos/grouped_mr_heart/demo_train.py
Here the training is launched using the GPU of index 0 with a limited number of steps
and reduced size. Please add flag --full to use the original training configuration,
such as
python demos/grouped_mr_heart/demo_train.py --full
Predict¶
Please execute the following command to run the prediction with pre-trained model. The
prediction logs and visualization results will be saved under
demos/grouped_mr_heart/logs_predict. Check the CLI documentation
for more details about prediction output.
python demos/grouped_mr_heart/demo_predict.py
Optionally, the user-trained model can be used by changing the ckpt_path variable
inside demo_predict.py. Note that the path should end with .ckpt and checkpoints are
saved under logs_train as mentioned above.
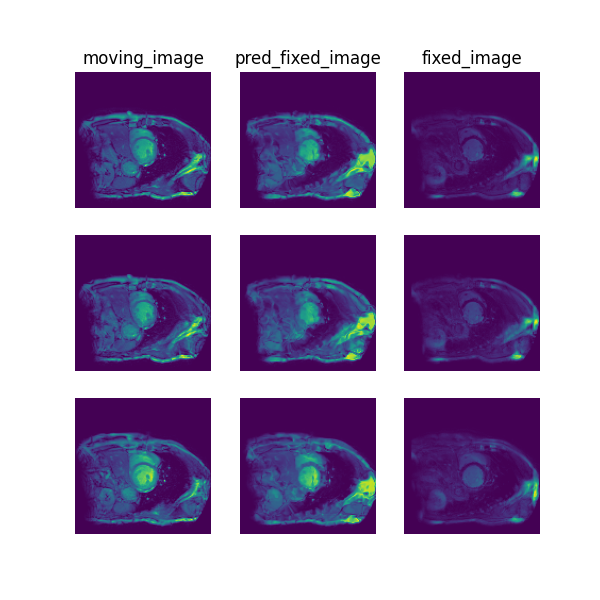
Visualise¶
The following command can be executed to generate a plot of three image slices from the the moving image, warped image and fixed image (left to right) to visualise the registration. Please see the visualisation tool docs here for more visualisation options such as animated gifs.
deepreg_vis -m 2 -i 'demos/grouped_mr_heart/logs_predict/<time-stamp>/test/<pair-number>/moving_image.nii.gz, demos/grouped_mr_heart/logs_predict/<time-stamp>/test/<pair-number>/pred_fixed_image.nii.gz, demos/grouped_mr_heart/logs_predict/<time-stamp>/test/<pair-number>/fixed_image.nii.gz' --slice-inds '14,10,20' -s demos/grouped_mr_heart/logs_predict
Note: The prediction must be run before running the command to generate the
visualisation. The <time-stamp> and <pair-number> must be entered by the user.
Contact¶
Please raise an issue for any questions.
Reference¶
[1] Xiahai Zhuang: Multivariate mixture model for myocardial segmentation combining multi-source images. IEEE Transactions on Pattern Analysis and Machine Intelligence (T PAMI), vol. 41, no. 12, 2933-2946, Dec 2019. link.
[2] Xiahai Zhuang: Multivariate mixture model for cardiac segmentation from multi-sequence MRI. International Conference on Medical Image Computing and Computer-Assisted Intervention, pp.581-588, 2016.