Pairwise registration for grouped prostate segmentation masks¶
Note: Please read the DeepReg Demo Disclaimer.
This demo uses DeepReg to demonstrate a number of features:
For grouped data in h5 files, e.g. “group-1-2” indicates the 2th visit from Subject 1;
Use masks as the images for feature-based registration - aligning the prostate gland segmentation in this case - with deep learning;
Register intra-patient longitudinal data.
This demo also implements the feature-based registration described in Morphological Change Forecasting for Prostate Glands using Feature-based Registration and Kernel Density Extrapolation.
Author¶
DeepReg Development Team
Application¶
Longitudinal registration detects the temporal changes and normalises the spatial difference between images acquired at different time-points. For prostate cancer patients under active surveillance programmes, quantifying these changes is useful for detecting and monitoring potential cancerous regions.
Data¶
This is a demo without real clinical data due to regulatory restrictions. The MR and ultrasound images used are simulated dummy 3D Ultrasound images. Data are organized into 10 separate folds.
Instruction¶
Installation¶
Please install DeepReg following the instructions and
change the current directory to the root directory of DeepReg project, i.e. DeepReg/.
Download data¶
Please execute the following command to download/pre-process the data and download the pre-trained model.
python demos/grouped_mask_prostate_longitudinal/demo_data.py
Launch demo training¶
Please execute the following command to launch a demo training (the first of the ten
runs of a 9-fold cross-validation). The training logs and model checkpoints will be
saved under demos/grouped_mask_prostate_longitudinal/logs_train.
python demos/grouped_mask_prostate_longitudinal/demo_train.py
Here the training is launched using the GPU of index 0 with a limited number of steps
and reduced size. Please add flag --full to use the original training configuration,
such as
python demos/grouped_mask_prostate_longitudinal/demo_train.py --full
Predict¶
Please execute the following command to run the prediction with pre-trained model. The
prediction logs and visualization results will be saved under
demos/grouped_mask_prostate_longitudinal/logs_predict. Check the
CLI documentation for more details about prediction output.
python demos/grouped_mask_prostate_longitudinal/demo_predict.py
Optionally, the user-trained model can be used by changing the ckpt_path variable
inside demo_predict.py. Note that the path should end with .ckpt and checkpoints are
saved under logs_train as mentioned above.
Visualise¶
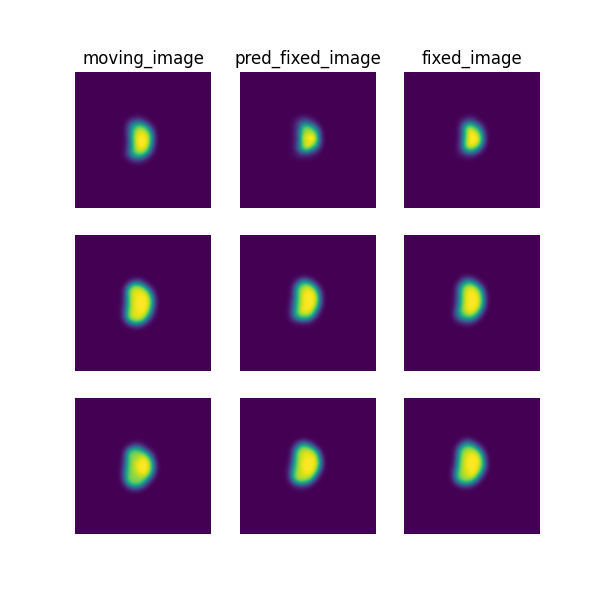
The following command can be executed to generate a plot of three image slices from the the moving image, warped image and fixed image (left to right) to visualise the registration. Please see the visualisation tool docs here for more visualisation options such as animated gifs.
deepreg_vis -m 2 -i 'demos/grouped_mask_prostate_longitudinal/logs_predict/<time-stamp>/test/<pair-number>/moving_image.nii.gz, demos/grouped_mask_prostate_longitudinal/logs_predict/<time-stamp>/test/<pair-number>/pred_fixed_image.nii.gz, demos/grouped_mask_prostate_longitudinal/logs_predict/<time-stamp>/test/<pair-number>/fixed_image.nii.gz' --slice-inds '10,16,20' -s demos/grouped_mask_prostate_longitudinal/logs_predict
Note: The prediction must be run before running the command to generate the
visualisation. The <time-stamp> and <pair-number> must be entered by the user.
Contact¶
Please raise an issue for any questions.