Classical affine registration for head-and-neck CT images¶
Note: Please read the DeepReg Demo Disclaimer.
This is a special demo that uses the DeepReg package for classical affine image registration, which iteratively solves an optimisation problem. Gradient descent is used to minimise the image dissimilarity function of a given pair of moving anf fixed images.
Author¶
DeepReg Development Team
Application¶
Although in this demo the moving images are simulated using a randomly generated transformation. The registration technique can be used in radiotherapy to compensate the difference between CT acquired at different time points, such as pre-treatment and intra-/post-treatment.
Data¶
Data is an example CT volume with two labels.
Instruction¶
Installation¶
Please install DeepReg following the instructions and
change the current directory to the root directory of DeepReg project, i.e. DeepReg/.
Download data¶
Please execute the following command to download and pre-process the data.
python demos/classical_ct_headneck_affine/demo_data.py
Launch registration¶
Please execute the following command to register two images. The fixed image will be the downloaded data and the moving image will be simulated by applying a random affine transformation, such that the ground-truth is available for. The optimised transformation will be applied to the moving images, as well as the moving labels. The results, saved in a timestamped folder under the project directory, will compare the warped image/labels with the ground-truth image/labels.
python demos/classical_ct_headneck_affine/demo_register.py
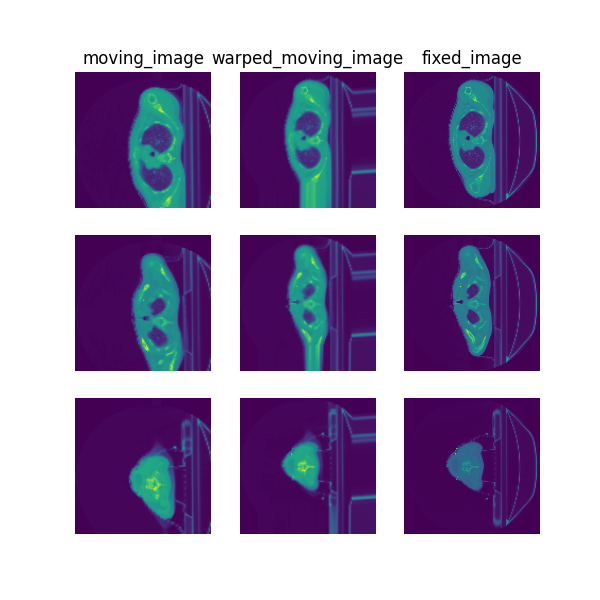
Visualise¶
The following command can be executed to generate a plot of three image slices from the the moving image, warped image and fixed image (left to right) to visualise the registration. Please see the visualisation tool docs here for more visualisation options such as animated gifs.
deepreg_vis -m 2 -i 'demos/classical_ct_headneck_affine/logs_reg/moving_image.nii.gz, demos/classical_ct_headneck_affine/logs_reg/warped_moving_image.nii.gz, demos/classical_ct_headneck_affine/logs_reg/fixed_image.nii.gz' --slice-inds '4,8,12' -s demos/classical_ct_headneck_affine/logs_reg
Note: The registration script must be run before running the command to generate the visualisation.
Contact¶
Please raise an issue for any questions.
Reference¶
[1] Vallières, M. et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 7, 10117 (2017). doi: 10.1038/s41598-017-10371-5