Unpaired hippocampus MR registration¶
Note: Please read the DeepReg Demo Disclaimer.
Author¶
DeepReg Development Team (Adrià Casamitjana)
Application¶
This is a demo targeting the alignment of hippocampal substructures (head and body) using mono-modal MR images between different patients. The images are cropped around those areas and manually annotated. This is a 3D intra-modal registration using a composite loss of image and label similarity.
Data¶
The dataset for this demo comes from the Learn2Reg MICCAI Challenge (Task 4) [1] and can be downloaded from: https://drive.google.com/uc?export=download&id=1RvJIjG2loU8uGkWzUuGjqVcGQW2RzNYA
Instruction¶
Installation¶
Please install DeepReg following the instructions and
change the current directory to the root directory of DeepReg project, i.e. DeepReg/.
Download data¶
Please execute the following command to download/pre-process the data and download the pre-trained model.
python demos/unpaired_mr_brain/demo_data.py
Pre-processing includes:
Rescaling all images’ intensity to 0-255.
Creating and applying a binary mask to mask-out the padded values in images.
Transforming label volumes using one-hot encoding (only for foreground classes)
Launch demo training¶
Please execute the following command to launch a demo training. The training logs and
model checkpoints will be saved under demos/unpaired_mr_brain/logs_train.
python demos/unpaired_mr_brain/demo_train.py
Here the training is launched using the GPU of index 0 with a limited number of steps
and reduced size. Please add flag --full to use the original training configuration,
such as
python demos/unpaired_mr_brain/demo_train.py --full
Predict¶
Please execute the following command to run the prediction with pre-trained model. The
prediction logs and visualization results will be saved under
demos/unpaired_mr_brain/logs_predict. Check the CLI documentation
for more details about prediction output.
python demos/unpaired_mr_brain/demo_predict.py
Visualise¶
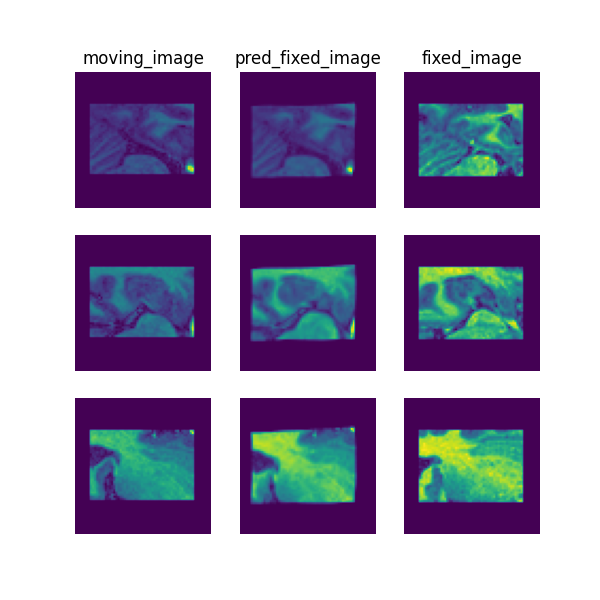
The following command can be executed to generate a plot of three image slices from the the moving image, warped image and fixed image (left to right) to visualise the registration. Please see the visualisation tool docs here for more visualisation options such as animated gifs.
deepreg_vis -m 2 -i 'demos/unpaired_mr_brain/logs_predict/<time-stamp>/test/<pair-number>/moving_image.nii.gz, demos/unpaired_mr_brain/logs_predict/<time-stamp>/test/<pair-number>/pred_fixed_image.nii.gz, demos/unpaired_mr_brain/logs_predict/<time-stamp>/test/<pair-number>/fixed_image.nii.gz' --slice-inds '20,32,44' -s demos/unpaired_mr_brain/logs_predict/
Note: The prediction must be run before running the command to generate the
visualisation. The <time-stamp> and <pair-number> must be entered by the user.
Contact¶
Please raise an issue for any questions.
Reference¶
[1] AL Simpson et al., A large annotated medical image dataset for the development and evaluation of segmentation algorithms (2019). https://arxiv.org/abs/1902.09063