Paired prostate MR-ultrasound registration¶
Note: Please read the DeepReg Demo Disclaimer.
This demo uses DeepReg to re-implement the algorithms described in Weakly-supervised convolutional neural networks for multimodal image registration. A standalone demo was hosted at https://github.com/yipenghu/label-reg.
Author¶
DeepReg Development Team
Application¶
Registering preoperative MR images to intraoperative transrectal ultrasound images has been an active research area for more than a decade. The multimodal image registration task assist a number of ultrasound-guided interventions and surgical procedures, such as targeted biopsy and focal therapy for prostate cancer patients. One of the key challenges in this registration task is the lack of robust and effective similarity measures between the two image types. This demo implements a weakly-supervised learning approach to learn voxel correspondence between intensity patterns between the multimodal data, driven by expert-defined anatomical landmarks, such as the prostate gland segmentaion.
Data¶
This is a demo without real clinical data due to regulatory restrictions. The MR and ultrasound images used are simulated dummy images.
Instruction¶
Installation¶
Please install DeepReg following the instructions and
change the current directory to the root directory of DeepReg project, i.e. DeepReg/.
Download data¶
Please execute the following command to download/pre-process the data and download the pre-trained model.
python demos/paired_mrus_prostate/demo_data.py
Launch demo training¶
Please execute the following command to launch a demo training (the first of the ten
runs of a 9-fold cross-validation). The training logs and model checkpoints will be
saved under demos/paired_mrus_prostate/logs_train.
python demos/paired_mrus_prostate/demo_train.py
Here the training is launched using the GPU of index 0 with a limited number of steps
and reduced size. Please add flag --full to use the original training configuration,
such as
python demos/paired_mrus_prostate/demo_train.py --full
Predict¶
Please execute the following command to run the prediction with pre-trained model. The
prediction logs and visualization results will be saved under
demos/paired_mrus_prostate/logs_predict. Check the
CLI documentation for more details about prediction output.
python demos/paired_mrus_prostate/demo_predict.py
Optionally, the user-trained model can be used by changing the ckpt_path variable
inside demo_predict.py. Note that the path should end with .ckpt and checkpoints are
saved under logs_train as mentioned above.
Visualise¶
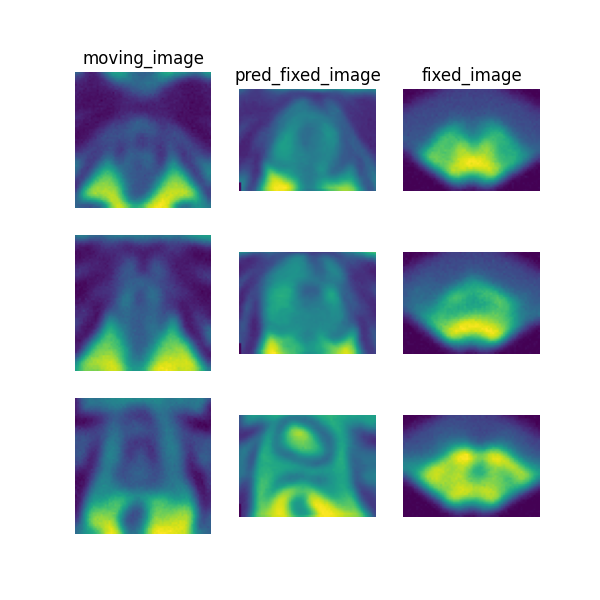
The following command can be executed to generate a plot of three image slices from the the moving image, warped image and fixed image (left to right) to visualise the registration. Please see the visualisation tool docs here for more visualisation options such as animated gifs.
deepreg_vis -m 2 -i 'demos/paired_mrus_prostate/logs_predict/<time-stamp>/test/<pair-number>/moving_image.nii.gz, demos/paired_mrus_prostate/logs_predict/<time-stamp>/test/<pair-number>/pred_fixed_image.nii.gz, demos/paired_mrus_prostate/logs_predict/<time-stamp>/test/<pair-number>/fixed_image.nii.gz' --slice-inds '12,20,36' -s demos/paired_mrus_prostate/logs_predict
Note: The prediction must be run before running the command to generate the
visualisation. The <time-stamp> and <pair-number> must be entered by the user.
Contact¶
Please raise an issue for any questions.